If you frequently deliver source code review assessments of products, including machine learning components, I’m sure you are used to reviewing Jupyter Notebook files (usually python). Although I spend most of my time reviewing the source code manually, I also use static analysis tools such as semgrep, using both public and private rules. This tool is handy if the source code is bigger than I can cover manually or finding certain types of vulnerabilities that makes much more sense using an automatic approach. If you haven’t used semgrep before, it is a powerful tool that can be used to detect vulnerabilities in the source code based on signatures called “rules”. I strongly recommend it.
However, I found that Semgrep was ignoring Jupyter Notebook files (extension .ipynb), so vulnerabilities included in those files remained undetected. This miss was because Jupyter Notebook files are not raw source code files but a JSON file format looking as follows:
{
"cell_type": "code",
"execution_count": 3,
"id": "28ed696c",
"metadata": {},
"outputs": [],
"source": [
"data = ["This", "is", "an", "example"] n",
"with open("test.pkl", 'wb') as f:n",
" pickle.dump(data, f)"
]
},For this reason, you will not detect a Jupyter Notebook loading a pickle-based serialised model or any other vulnerability introduced in these files. There is a workaround, which is using the “jupyter” command line utility to convert the notebook file into a raw python file, as shown below:
$ jupyter nbconvert --to python test.ipynbIf you combine this with a bit of shell scripting, you can create an equivalent .py file for every .ipynb file and run Semgrep through those files.
But is there any way to detect them using Semgrep’s built-in features? The answer is “almost”. An experimental feature called “extract mode” (documentation here) was designed for a very similar situation. With the extract mode, you can create a rule to extract specific content from a file format and process it as code for a different language. So, for example, a shell command can be extracted from RUN’s Dockerfile command and processed as shell code instead of Dockerfile.
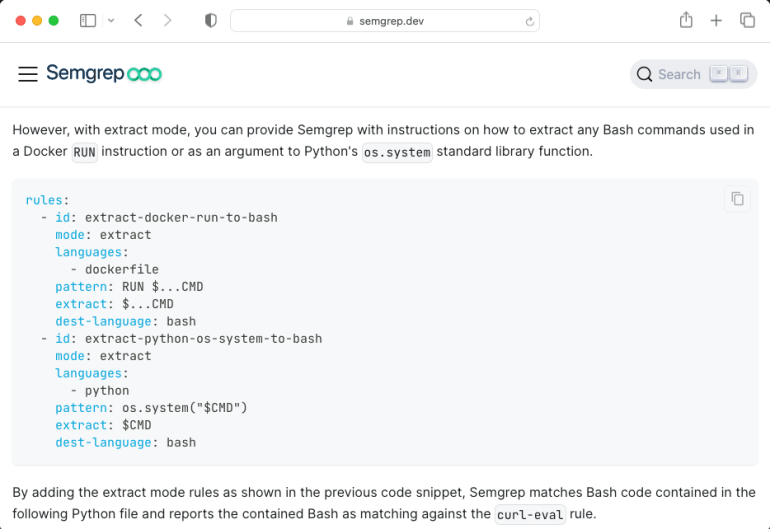
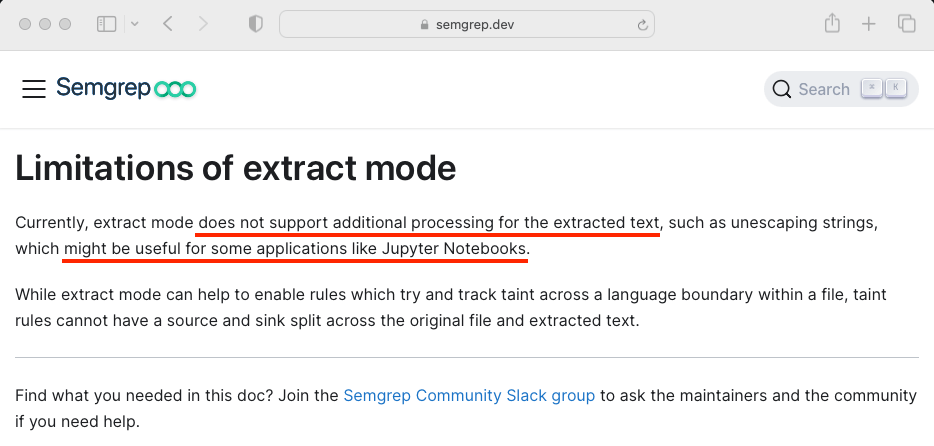
The problem with this fantastic feature is that it extracts raw source code, but it was not designed to process the extracted content, as mentioned in the documentation.
For this reason, we decided to contribute this feature to Semgrep via this pull request (GitHub link here). Now the extract mode has a new flag called “transform”, specifying if the extracted content should be processed as raw source code (default) or JSON array (concat_json_string_array). As a result, we can now create an extraction rule as follows.
rules:
- id: extract-jupyter-to-python
mode: extract
languages:
- json
pattern: |
"source": $CODE
extract: $CODE
reduce: concat
transform: concat_json_string_array
dest-language: pythonFinally, we can run our semgrep command, including our extraction rule for Jupyter Notebook, so those files are also covered.
$ semgrep -c extract-jupyter-to-python.yaml -c "p/security-audit" src/
Semgrep rule registry URL is https://semgrep.dev/registry.
Scanning across multiple languages:
| 3 rules × 1 file
json | 2 rules × 1 file
Findings:
src/test.ipynb
python.lang.security.deserialization.pickle.avoid-pickle
Avoid using `pickle`, which is known to lead to code execution vulnerabilities. When
unpickling, the serialized data could be manipulated to run arbitrary code. Instead,
consider serializing the relevant data as JSON or a similar text-based serialization format.
Details: https://sg.run/xNoj
30┆ " pickle.dump(data, f)n", It is worth mentioning that extract mode is an experimental feature and it could be modified in the future. So please keep that in mind before using it in your pipeline.Any issue that you find in this new experimental feature, feel free to report it in GitHub as an issue.
